BioGateway Use Case Step-by-step CREB-CREM example
This section describes how the information from ExTRI is used and produced using the compiled TF-TG graph in the BioGateway triple store. To reproduce this results, the reader is advised to look at the tutorial published in the Current Protocols BioGateway App Use Case paper.
Define Settings panel of BioGateway App
In the Cytoscape network editor, install the Biogateway App from the App store or from the BioGateway website, and launch the App. Before preparing a query, the settings need to be specified, as shown in Figure 1.
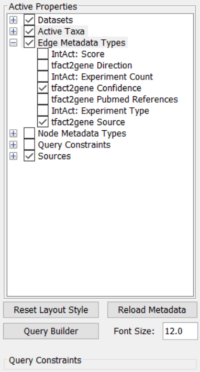
Figure 1: Settings panel BioGateway App
| Datasets: | all |
| Active taxa: | Homo sapiens |
| Edge metadata: | tfact2gene confidence |
| tfact2gene source | |
| Node metadata: | none |
| Query constraints: | none |
| Sources (TF-TG): | all |
Building the Query in BioGateway
Next, launch the Query Builder and define the following query (Figure 2).
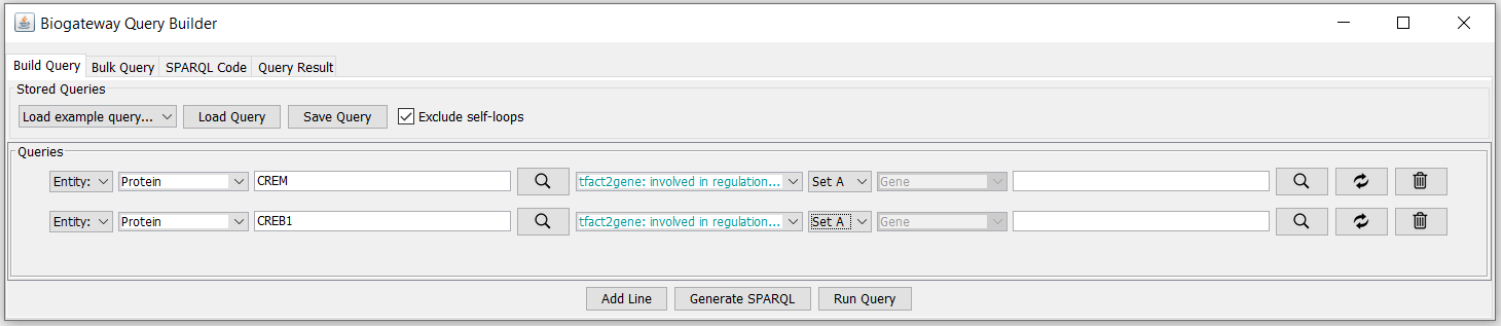
Figure 2: Screenshot of the query builder.
This query will search for all genes from the tfact2gene graph that are regulated by the transcription factor (protein) CREM, and next, all genes regulated transcription factor CREB1. As the genes are defined as having to be in the same set, the query constrains the genes to those connected to both TFs. Run Query produces the results in Figure 3.
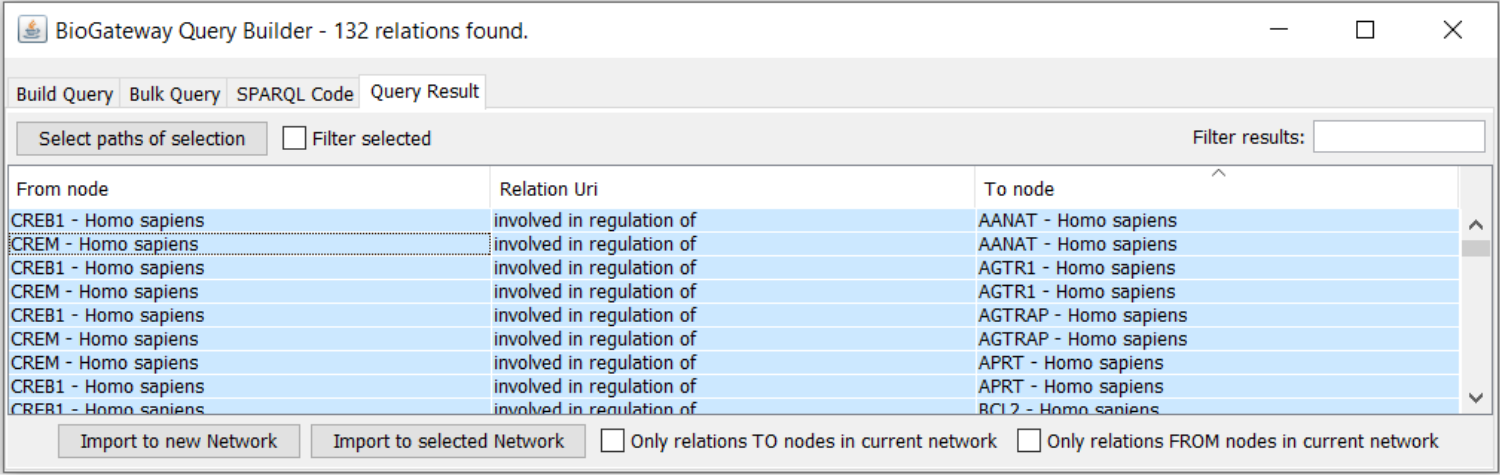
Figure 3: The results from the query.
Importing the Query into a new network
By right-click and Ctr+A, the results are highlighted for Import to New Network. The network imported to the Cytoscape canvas contains the edge metadata (Figure 1) specifying the level of Confidence associated with the TRI, and the Source of the TRI, as shown in the right-most section of the Edge Table (figure 4). Sorting for the confidence level by clicking on the column header groups all TRI for confidence, and after highlighting all relationships that do not have ‘High’ as Confidence these TRIs can be deleted from the network.
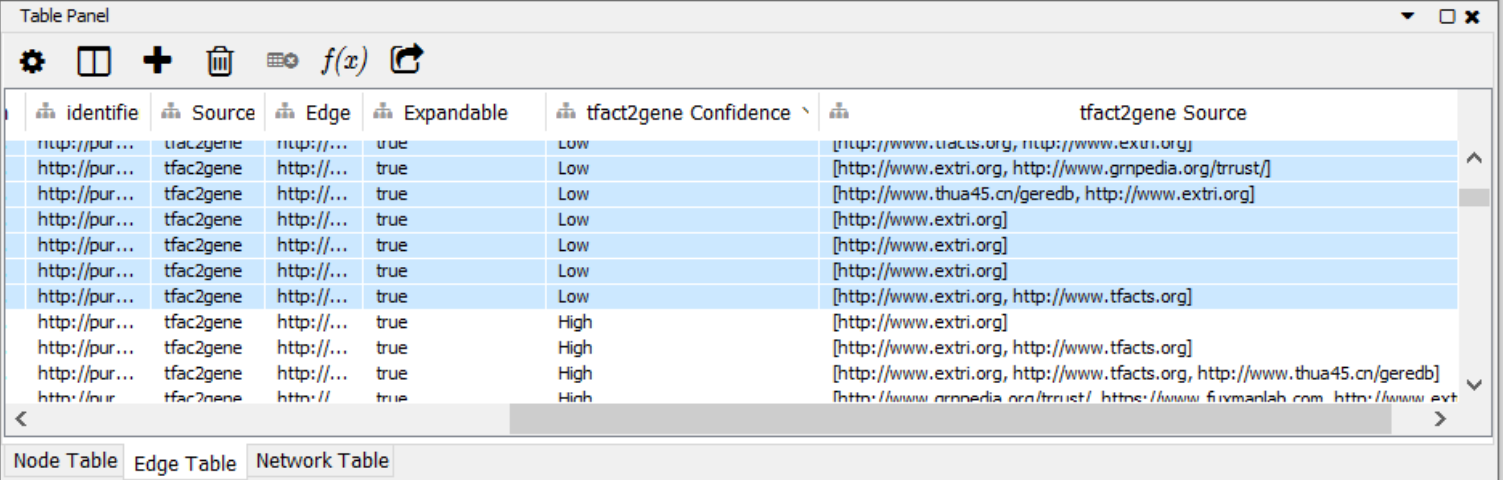
Figure 4: Edge table with Confidence and Source columns of the TRI.
The relation between CREM and CRH
The remaining network links (green edges) the two TFs CREM and CREB1 (light blue boxes) to 41 genes (yellow ovals), representing 82 TRI (Figure 5). This network was rearranged to display all genes that were part of at least one TRI uniquely present in the ExTRI corpus in a top row with seven genes (showing CRH, PER1, IL17A, FSHB, AGTR1, IL4, PTH) above the TFs, and all other genes not uniquely dependent on ExTRI in the cluster below that. Double clicking on the relation between CREM and CRH (green arrow, left panel Figure 5) expands the link and produces a node that provides access to the landing page (round node in the right panel of figure 5, between CREM and CRG).
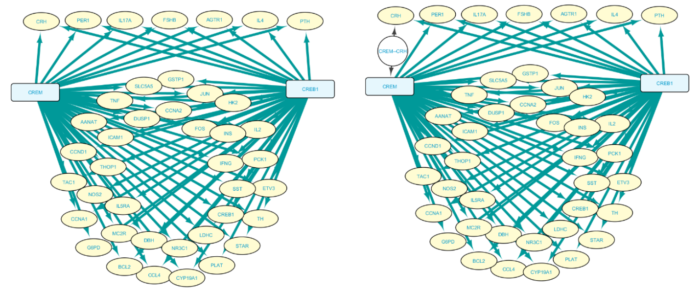
Figure 5: TF-TG network pruned for High-confidence TRIs. The right panel has an expanded relationship (round node) between TF: CREM and TG: CRH.
From network edges to landing pages
Right clicking on the expanded relationship node pops up a menu option provided by the App (Figure 6): BioGateway > Open resource URI, which will open the landing page for this TF/TG relationship in a new window.
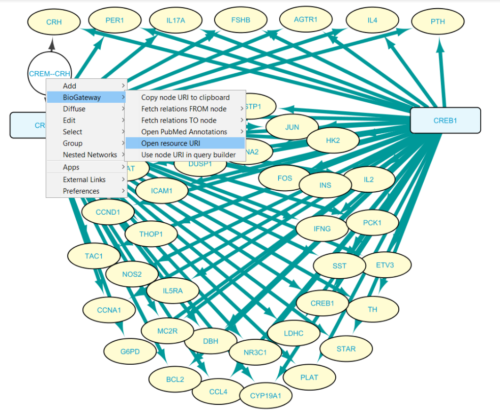
Figure 6: From network edges to landing pages.
The landing page (Figure 7) shows the source of the TRI (multiple sources are possible), and if the source includes ExTRI, an image of the abstract is produced, with the TRI sentence highlighted: